-
PDF
- Split View
-
Views
-
Cite
Cite
Yousong Peng, Lei Yang, Honglei Li, Yuanqiang Zou, Lizong Deng, Aiping Wu, Xiangjun Du, Dayan Wang, Yuelong Shu, Taijiao Jiang, PREDAC-H3: a user-friendly platform for antigenic surveillance of human influenza a(H3N2) virus based on hemagglutinin sequences, Bioinformatics, Volume 32, Issue 16, August 2016, Pages 2526–2527, https://doi.org/10.1093/bioinformatics/btw185
Close - Share Icon Share
Abstract
Motivation: Timely surveillance of the antigenic dynamics of the influenza virus is critical for accurate selection of vaccine strains, which is important for effective prevention of viral spread and infection.
Results: Here, we provide a computational platform, called PREDAC-H3, for antigenic surveillance of human influenza A(H3N2) virus based on the sequence of surface protein hemagglutinin (HA). PREDAC-H3 not only determines the antigenic variants and antigenic cluster (grouped for similar antigenicity) to which the virus belongs, based on HA sequences, but also allows visualization of the spatial distribution and temporal dynamics of antigenic clusters of viruses isolated from around the world, thus assisting in antigenic surveillance of human influenza A(H3N2) virus.
Availability and Implementation: It is publicly available from: http://biocloud.hnu.edu.cn/influ411/html/index.php.
Contacts: yshu@cnic.org.cn or taijiao@moon.ibp.ac.cn
1 Introduction
The seasonal influenza viruses, i.e. the influenza A(H1N1), A(H3N2) and B virus, causes continual epidemics to human society due to their frequent antigenic drift (Smith et al., 2004; Taubenberger and Kash, 2010). Timely surveillance of the antigenic dynamics of influenza viruses is critical for selection of appropriate vaccine strains. The World Health Organization (WHO) established a Global Influenza Surveillance Network, which is composed of National Influenza Centers from all over the world, to monitor the spread and antigenic variation of influenza viruses (WHO, 2014). Despite these global efforts, vaccine mismatches occasionally happen (Du et al., 2012; Salzberg, 2008).
Given the feasibility of rapid and high-quality sequence determination, a number of sequence-based computational approaches have been developed to understand the antigenic properties and characteristics of influenza virus evolution (Du et al., 2012; Lee and Chen, 2004; Luksza and Lassig, 2014; Peng et al., 2014). Previously, we developed a computational method PREDAC to infer antigenic clusters for human influenza A(H3N2) virus based on protein sequences of the immunodominant part of HA (HA1) (Du et al., 2012). By accurate capture of the antigenic characteristics of the virus, PREDAC proved helpful to improve the accuracy of vaccine recommendation in mainland China. Here, we made it easy to use for the public and further built a user-friendly platform PREDAC-H3 for antigenic surveillance of human influenza A(H3N2) virus. This could assist in timely and accurate selection of vaccine strains.
2 The PREDAC-H3 web server
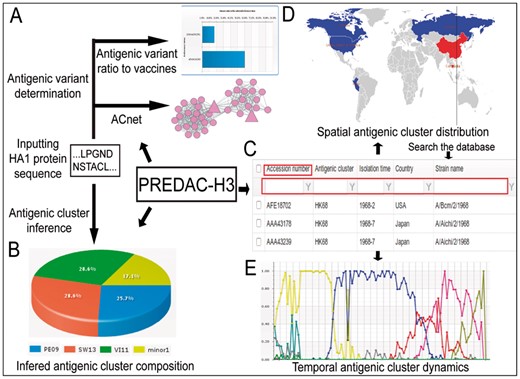
PREDAC-H3 has functionalities including determining the antigenic variant and antigenic cluster based on HA1 protein sequence, and displaying the spatial distribution and temporal dynamics of antigenic clusters in globe for human influenza A(H3N2) virus (Fig. 1). The functionalities of antigenic variant determination and antigenic cluster inference both take the HA1 protein sequences of the virus as inputs. The former predicts whether the viruses inputted are of similar or different antigenicity to previously recommended vaccine strains (Fig. 1A). An antigenic correlation network (ACnet) visualized using the flash version of Cytoscape (Shannon et al., 2003) is also provided, which neatly illustrates the antigenic relationships between the represented viruses (Fig. 1A). The latter infers the antigenic clusters to which the viruses inputted belong. A pie chart would be shown which display the composition of antigenic clusters for the viruses inputted (Fig. 1B).
Overview of PREDAC-H3. (A, B, D, E) The outputs for the functionalities of antigenic variant determination, antigenic cluster inference, spatial distribution and temporal dynamics of antigenic clusters, respectively. (C) The database included in the platform, which contains the isolation time, country, accession number and strain name of human influenza A(H3N2) viruses derived from Influenza Virus Resource and the predicted antigenic clusters to which they belong
The functionalities of displaying the spatial distribution and temporal dynamics of antigenic clusters for human influenza A(H3N2) virus rely on the determination of antigenic clusters for all the human influenza A(H3N2) viruses with HA1 protein sequence available in the database of Influenza Virus Resource (Bao et al., 2008), which would be conducted by PREDAC. The predicted antigenic clusters to which these viruses belong, and other (meta)data of the entries, including their time and country of isolation, accession number and strain name, are stored in a database within the platform (Fig. 1C). Users can search the database for the antigenic cluster to which their virus of interest belongs. In order to remain up-to-date and ensure that influenza A(H3N2) sequences recently submitted to the Influenza Virus Resource are included, the database is automatically updated on a monthly basis.
Based on the database, the platform can project the antigenic clusters circulating in the world for a selected month on a map (Fig. 1D). This is achieved by coloring the country or region in the represented map according to the dominant antigenic cluster (with the largest ratio) circulating in that region in a given month. When no virus sequences are available in the database in the given month, the location is presented in gray. Moreover, the temporal antigenic dynamics in a country or region is graphically represented as a further function of PREDAC-H3 (Fig. 1E). Each line in the chart represents the dynamic change of the ratio of an antigenic cluster circulating in a country or region over time. The coloring is performed by antigenic cluster, similar to the colors used to represent spatial dominance of antigenic clusters. Users can select the country or region and time period of interest to view the antigenic dynamics of the human influenza A(H3N2) virus.
Funding
This work has been supported the National Natural Science Foundation (31371338 and 31500126), Important National Science & Technology Specific Projects (2013ZX10004611-002 and 2014ZX10004002-001) and the International Scientific and Technological Cooperation project (2014DFB30010).
Conflict of Interest: none declared.
References
Author notes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
Associate Editor: John Hancock